Secondary_outcomes
Secondary_outcomes.RmdLoad data
#library(librarian)
#librarian::shelf(here, readxl, metafor, quiet = TRUE)
dat.second <- readxl::read_excel(here::here("data", "Dex_metan.xlsx"),
range = "B17:K79",
col_names = c("study",
"m1i", "sd1i",
"m2i", "sd2i",
"n1i", "n2i",
"summary1", "summary2",
"outcome")
)Secondary Outcome Analysis
Plateau pressure
dat.plat <- metafor::escalc(measure="MD",
m1i=m1i,
sd1i=sd1i,
n1i=n1i,
m2i=m2i,
sd2i=sd2i,
n2i=n2i,
data=dplyr::filter(dat.second, outcome=="Plateau Pressure"),
slab = study)
pm.plat <- metafor::rma(yi, vi, data=dat.plat, method="PM")
metafor::forest(pm.plat,
at=seq(from=-8, to =2, by=2))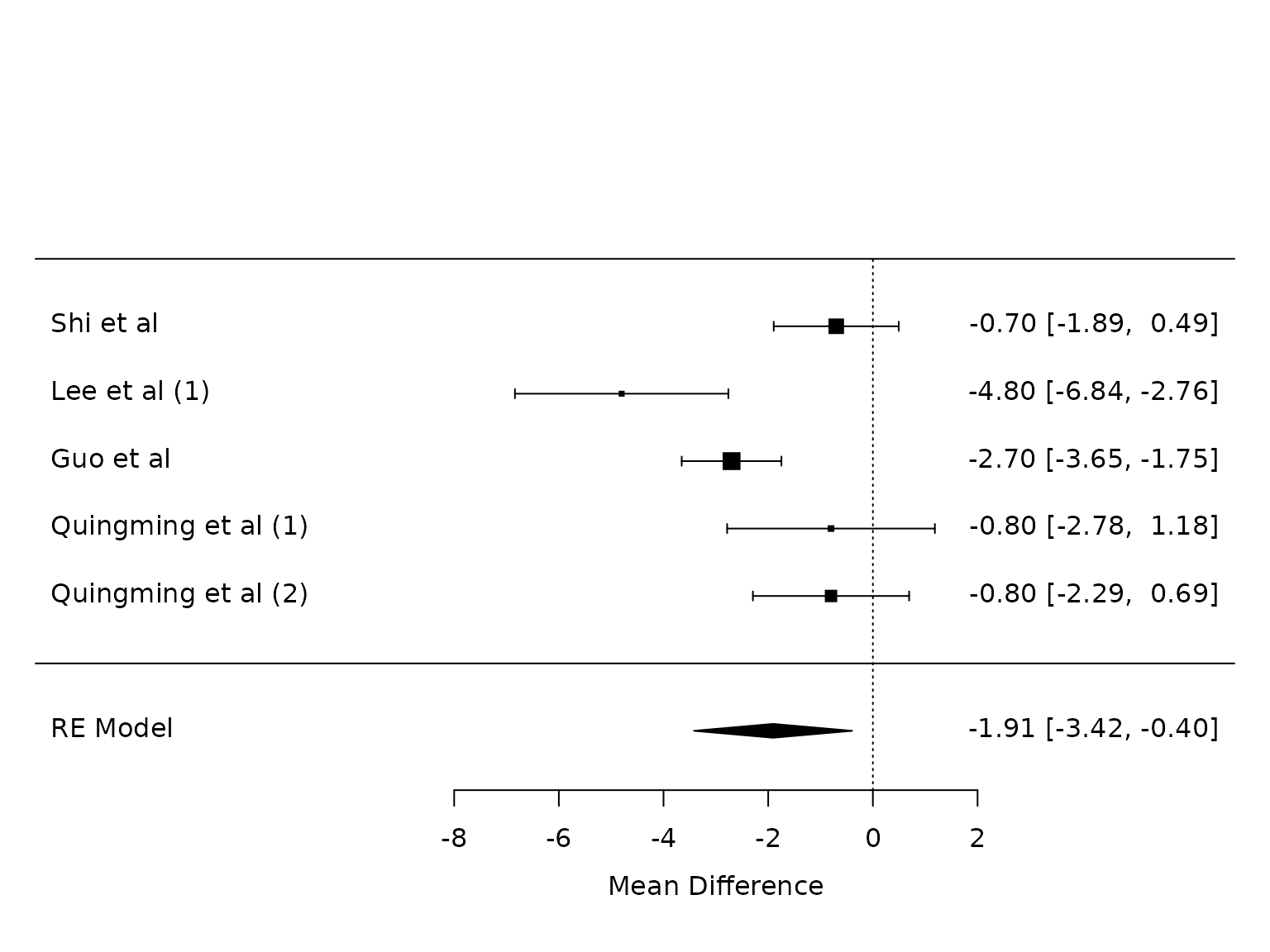
Peak pressure
dat.peak <- metafor::escalc(measure="MD",
m1i=m1i,
sd1i=sd1i,
n1i=n1i,
m2i=m2i,
sd2i=sd2i,
n2i=n2i,
data=dplyr::filter(dat.second, outcome=="Peak Pressure"),
slab = study)
pm.peak <- metafor::rma(yi, vi, data=dat.peak, method="PM")
metafor::forest(pm.peak,
at=seq(from=-8, to =2, by=2))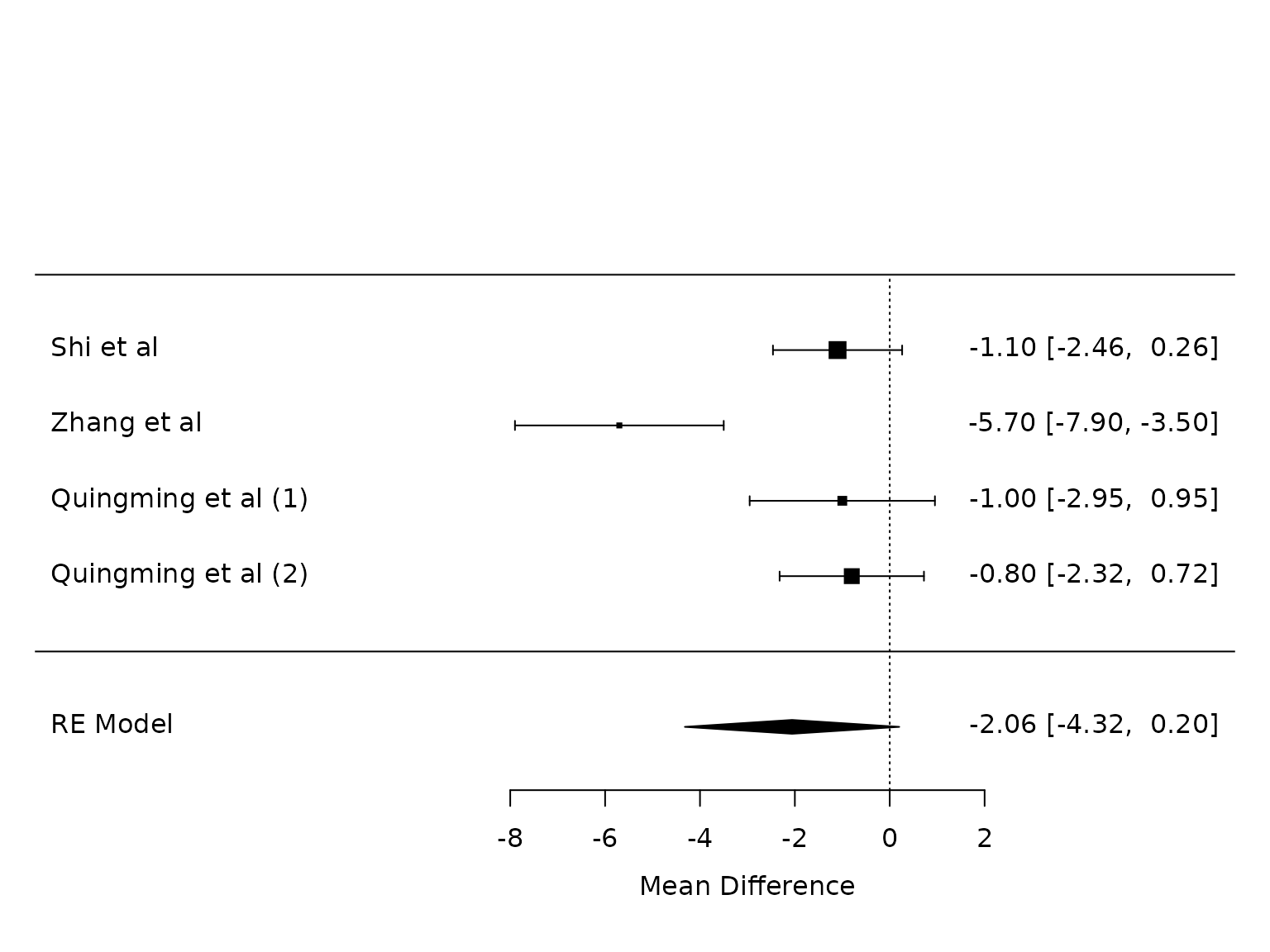
Respiratory Compliance:
dat.resp <- metafor::escalc(measure="MD",
m1i=m1i,
sd1i=sd1i,
n1i=n1i,
m2i=m2i,
sd2i=sd2i,
n2i=n2i,
data=dplyr::filter(dat.second, outcome=="Respiratory Compliance"),
slab = study)
pm.resp <- metafor::rma(yi, vi, data=dat.resp, method="PM")
metafor::forest(pm.resp,
at=seq(from=0, to =8, by=2))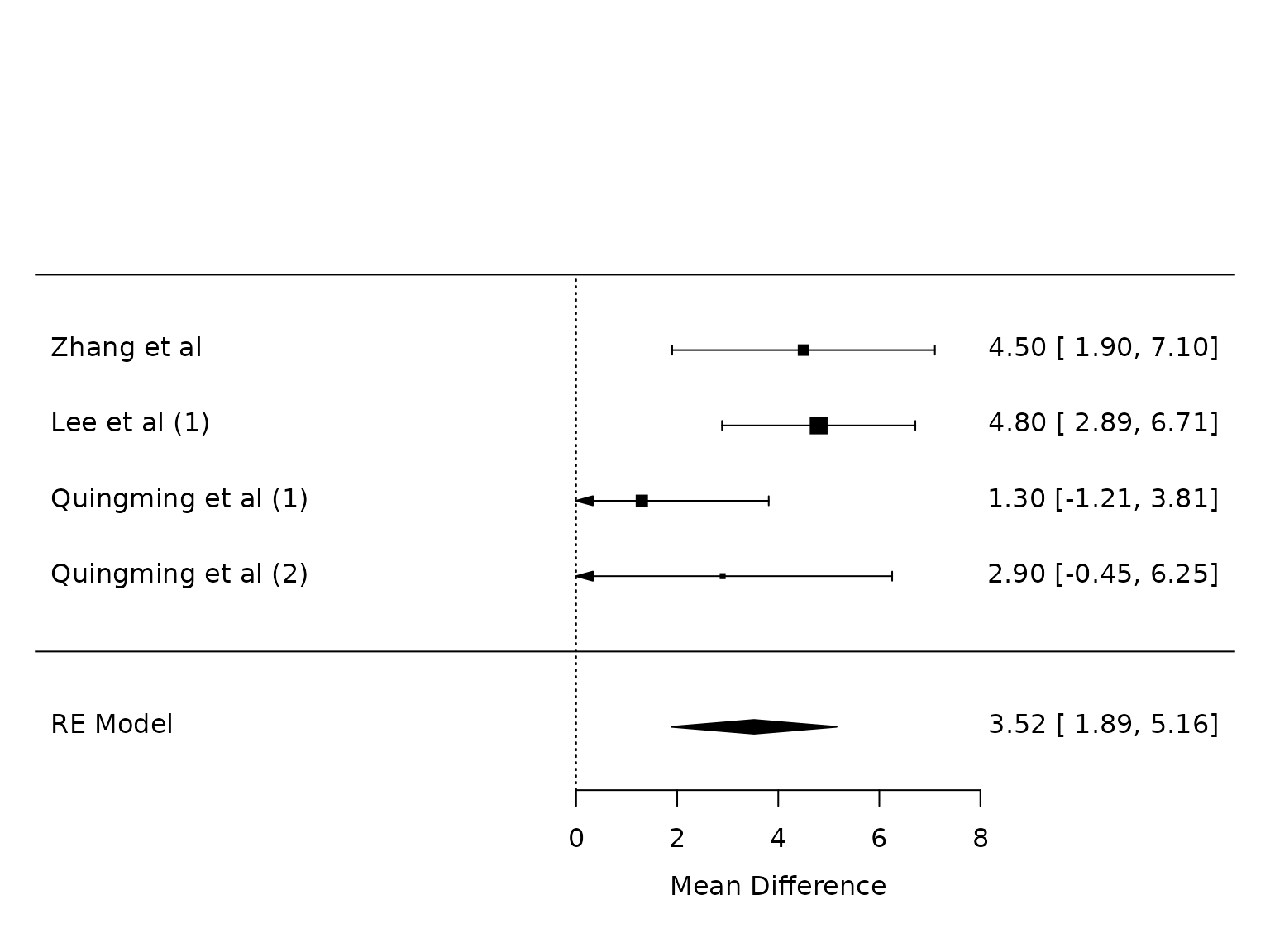
FEV1
dat.fev1<- metafor::escalc(measure="MD",
m1i=m1i,
sd1i=sd1i,
n1i=n1i,
m2i=m2i,
sd2i=sd2i,
n2i=n2i,
data=dat.second[grep(pattern = "FEV1", x = dat.second$outcome), ],
slab = study)
pm.fev1 <- metafor::rma(yi, vi, data=dat.fev1, method="PM")
metafor::forest(pm.fev1)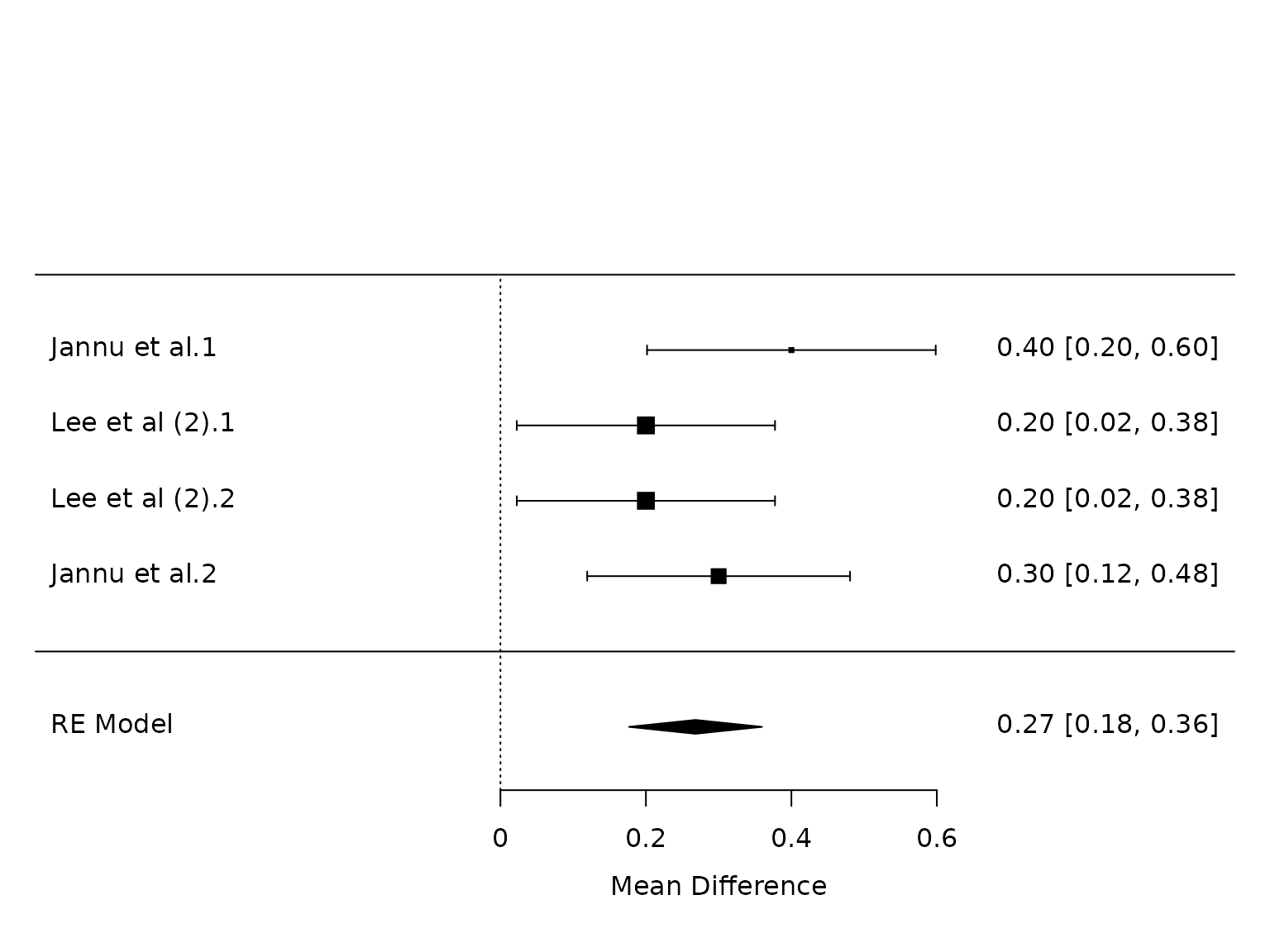
POD 1
dat.pod1<- metafor::escalc(measure="MD",
m1i=m1i,
sd1i=sd1i,
n1i=n1i,
m2i=m2i,
sd2i=sd2i,
n2i=n2i,
data=dplyr::filter(dat.second, outcome=="FEV1 POD 1"),
slab = study)
pm.pod1 <- metafor::rma(yi, vi, data=dat.pod1, method="PM")
metafor::forest(pm.pod1)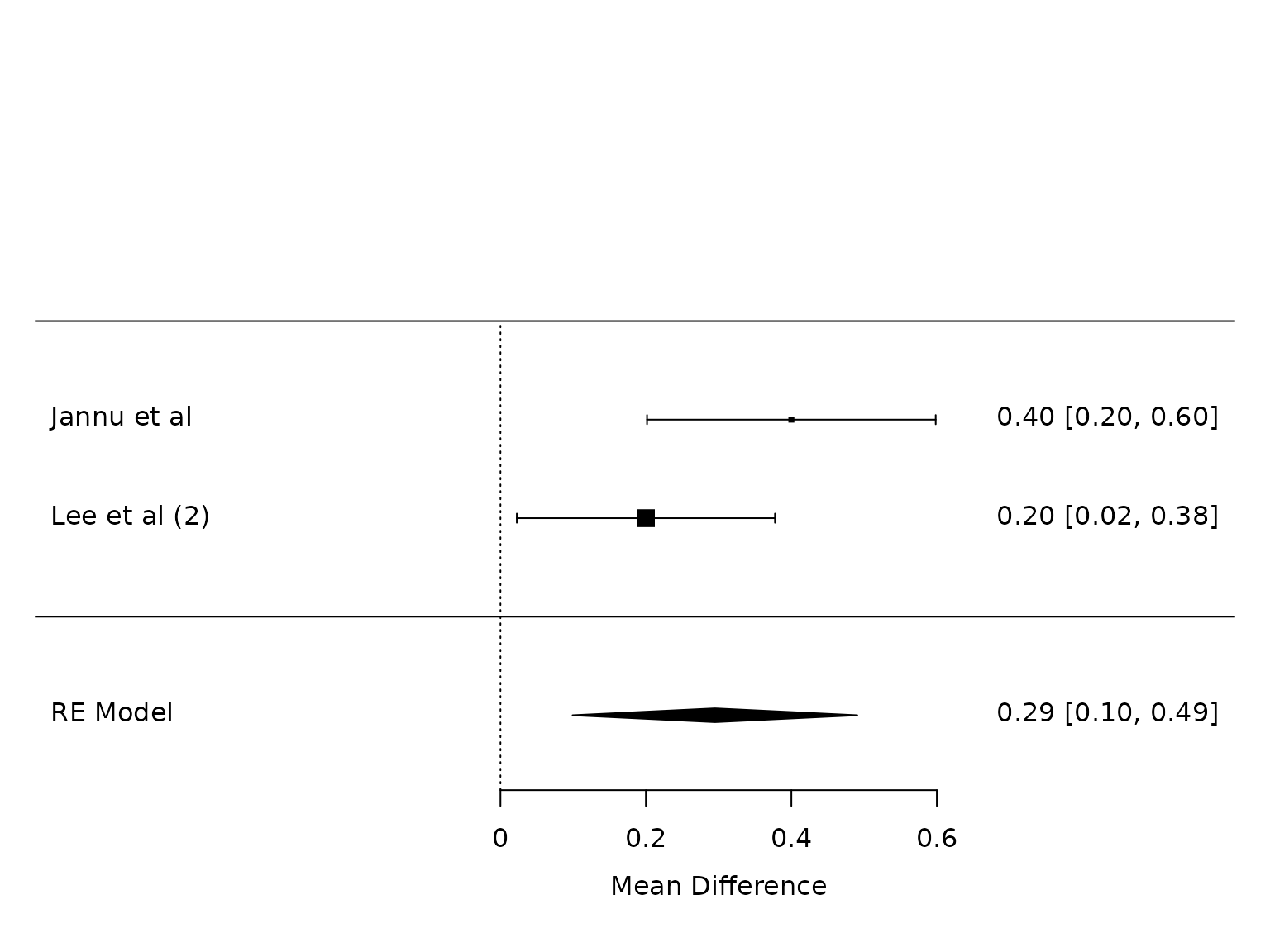
POD 2
dat.pod2<- metafor::escalc(measure="MD",
m1i=m1i,
sd1i=sd1i,
n1i=n1i,
m2i=m2i,
sd2i=sd2i,
n2i=n2i,
data=dplyr::filter(dat.second, outcome=="FEV1 POD 2"),
slab = study)
dat.pod2
#>
#> study m1i sd1i m2i sd2i n1i n2i summary1 summary2 outcome yi
#> 1 Lee et al (2) 2.2 0.5 2 0.4 50 50 <NA> <NA> FEV1 POD 2 0.2000
#> 2 Jannu et al 2.3 0.5 2 0.3 40 40 <NA> <NA> FEV1 POD 2 0.3000
#> vi
#> 1 0.0082
#> 2 0.0085
pm.pod2 <- metafor::rma(yi, vi, data=dat.pod2, method="PM")
weights(pm.pod2)
#> Lee et al (2) Jannu et al
#> 50.8982 49.1018
dat.pod2
#>
#> study m1i sd1i m2i sd2i n1i n2i summary1 summary2 outcome yi
#> 1 Lee et al (2) 2.2 0.5 2 0.4 50 50 <NA> <NA> FEV1 POD 2 0.2000
#> 2 Jannu et al 2.3 0.5 2 0.3 40 40 <NA> <NA> FEV1 POD 2 0.3000
#> vi
#> 1 0.0082
#> 2 0.0085
metafor::forest(pm.pod2)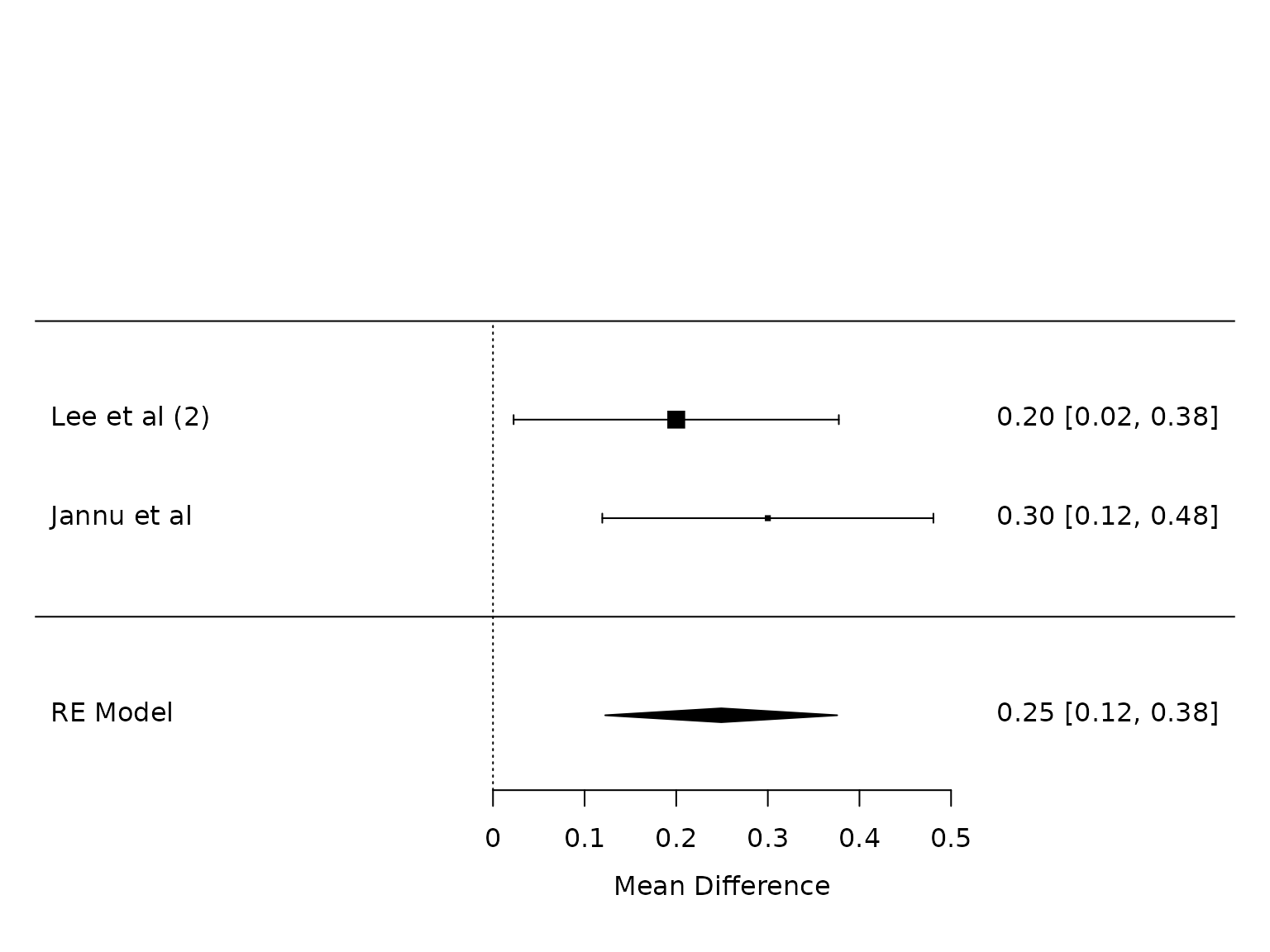
Length of Stay
dat.los <- metafor::escalc(measure="MD",
m1i=m1i,
sd1i=sd1i,
n1i=n1i,
m2i=m2i,
sd2i=sd2i,
n2i=n2i,
data=dplyr::filter(dat.second, outcome=="LOS"),
slab = study)
pm.los <- metafor::rma(yi, vi, data=dat.los, method="PM")
dat.los
#>
#> study m1i sd1i m2i sd2i n1i n2i summary1 summary2 outcome yi
#> 1 Meng et al 4.10 1.48 4.60 1.88 20 20 <NA> <NA> LOS -0.5000
#> 2 Zhang et al 7.10 3.90 7.76 3.12 28 28 <NA> <NA> LOS -0.6600
#> 3 Wu et al 5.60 2.50 5.90 2.50 30 30 <NA> <NA> LOS -0.3000
#> 4 Jannu et al 6.33 4.61 9.66 5.38 40 40 <NA> <NA> LOS -3.3300
#> 5 Lee et al (1) 7.16 3.93 8.03 3.93 25 25 <NA> <NA> LOS -0.8700
#> 6 Lee et al (2) 6.10 4.58 7.13 3.81 50 50 <NA> <NA> LOS -1.0300
#> vi
#> 1 0.2862
#> 2 0.8909
#> 3 0.4167
#> 4 1.2549
#> 5 1.2356
#> 6 0.7098
metafor::forest(pm.los)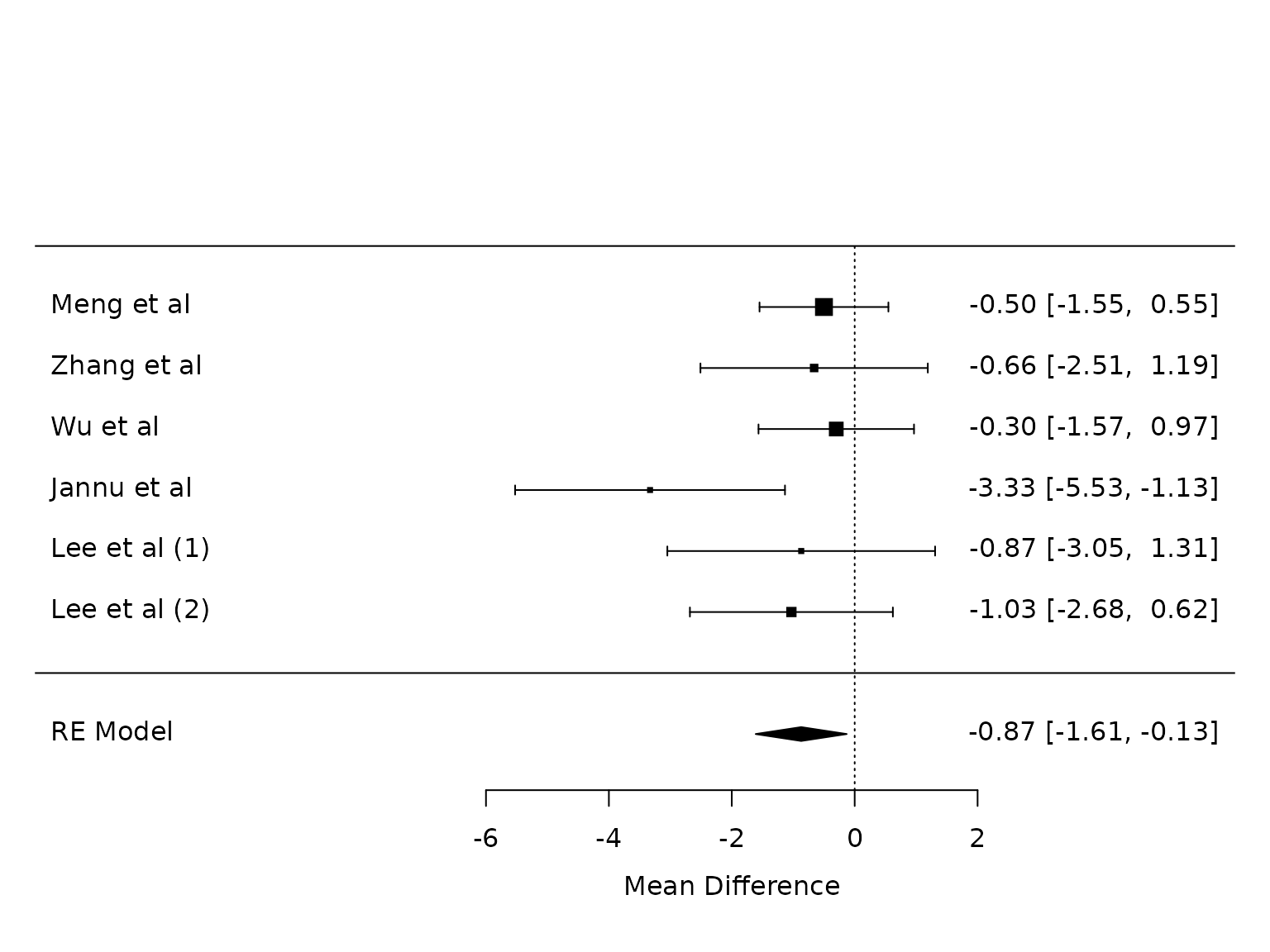
Excluding outlier Jannu et al.:
pm.los.exc.jannu <- metafor::leave1out(pm.los)
metafor::forest(pm.los.exc.jannu$estimate, sei=pm.los.exc.jannu$se, header=TRUE, xlab="Leave One Out Estimate", refline=coef(pm.los))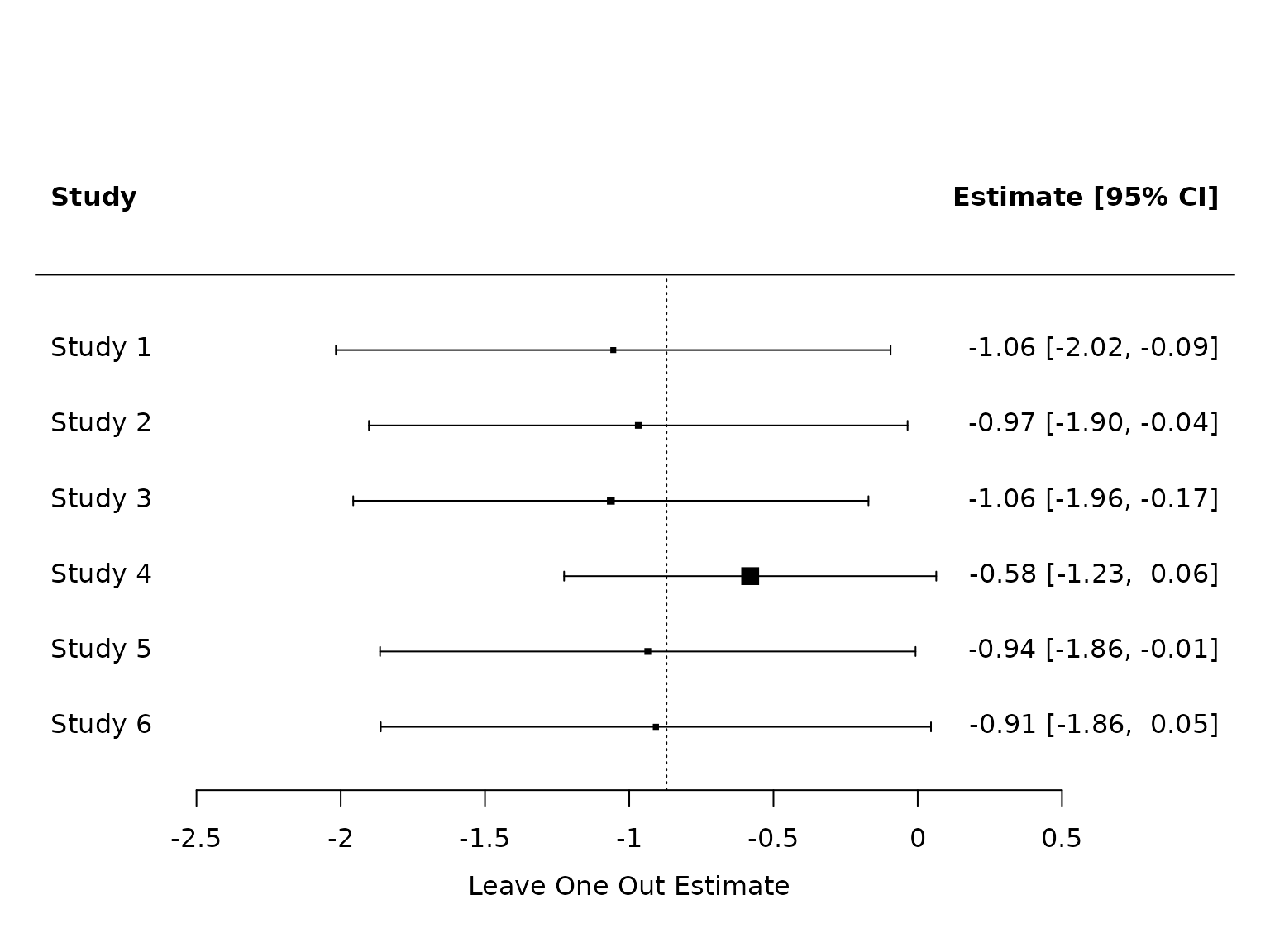
Note in the above figure that Study 4 is Jannu et al., hence this row gives the summary estimate when Jannu et al. is excluded.